Python client and API for accessing PIASOmarkerDB#
Import PIASO#
[1]:
import piaso
/n/data1/hms/neurobio/fishell/mindai/.conda/envs/nca/lib/python3.10/site-packages/tqdm/auto.py:21: TqdmWarning: IProgress not found. Please update jupyter and ipywidgets. See https://ipywidgets.readthedocs.io/en/stable/user_install.html
from .autonotebook import tqdm as notebook_tqdm
Import other packages#
[2]:
import numpy as np
import pandas as pd
import scanpy as sc
sc.set_figure_params(dpi=80,dpi_save=300, color_map='viridis',facecolor='white')
from matplotlib import rcParams
# To modify the default figure size, use rcParams.
rcParams['figure.figsize'] = 4, 4
rcParams['font.sans-serif'] = "Arial"
rcParams['font.family'] = "Arial"
sc.settings.verbosity = 3
sc.logging.print_header()
/tmp/ipykernel_4126893/1353975569.py:11: RuntimeWarning: Failed to import dependencies for application/vnd.jupyter.widget-view+json representation. (ModuleNotFoundError: No module named 'ipywidgets')
sc.logging.print_header()
[2]:
| Component | Info |
|---|---|
| Python | 3.10.19 (main, Oct 21 2025, 16:43:05) [GCC 11.2.0] |
| OS | Linux-5.14.0-570.23.1.el9_6.x86_64-x86_64-with-glibc2.34 |
| CPU | 32 logical CPU cores, x86_64 |
| GPU | No GPU found |
| Updated | 2026-01-20 19:17 |
Dependencies
| Dependency | Version |
|---|---|
| h5py | 3.15.1 |
| tqdm | 4.67.1 |
| stack_data | 0.6.3 |
| wcwidth | 0.2.13 |
| pillow | 12.0.0 |
| joblib | 1.5.3 |
| requests | 2.32.5 |
| debugpy | 1.8.16 |
| texttable | 1.7.0 |
| kiwisolver | 1.4.9 |
| asttokens | 3.0.0 |
| six | 1.17.0 |
| natsort | 8.4.0 |
| pure_eval | 0.2.3 |
| charset-normalizer | 3.4.4 |
| python-dateutil | 2.9.0.post0 |
| torch | 2.9.1 (2.9.1+cu128) |
| tornado | 6.5.4 |
| parso | 0.8.5 |
| pytz | 2025.2 |
| leidenalg | 0.10.2 |
| jedi | 0.19.2 |
| networkx | 3.4.2 |
| executing | 2.2.1 |
| setuptools | 80.9.0 |
| cycler | 0.12.1 |
| igraph | 0.11.9 |
| statsmodels | 0.14.6 |
| ipython | 8.30.0 |
| decorator | 5.2.1 |
| certifi | 2026.1.4 (2026.01.04) |
| patsy | 1.0.2 |
| psutil | 7.0.0 |
| llvmlite | 0.46.0 |
| prompt_toolkit | 3.0.52 |
| numba | 0.63.1 |
Copyable Markdown
| Dependency | Version | | ------------------ | --------------------- | | h5py | 3.15.1 | | tqdm | 4.67.1 | | stack_data | 0.6.3 | | wcwidth | 0.2.13 | | pillow | 12.0.0 | | joblib | 1.5.3 | | requests | 2.32.5 | | debugpy | 1.8.16 | | texttable | 1.7.0 | | kiwisolver | 1.4.9 | | asttokens | 3.0.0 | | six | 1.17.0 | | natsort | 8.4.0 | | pure_eval | 0.2.3 | | charset-normalizer | 3.4.4 | | python-dateutil | 2.9.0.post0 | | torch | 2.9.1 (2.9.1+cu128) | | tornado | 6.5.4 | | parso | 0.8.5 | | pytz | 2025.2 | | leidenalg | 0.10.2 | | jedi | 0.19.2 | | networkx | 3.4.2 | | executing | 2.2.1 | | setuptools | 80.9.0 | | cycler | 0.12.1 | | igraph | 0.11.9 | | statsmodels | 0.14.6 | | ipython | 8.30.0 | | decorator | 5.2.1 | | certifi | 2026.1.4 (2026.01.04) | | patsy | 1.0.2 | | psutil | 7.0.0 | | llvmlite | 0.46.0 | | prompt_toolkit | 3.0.52 | | numba | 0.63.1 | | Component | Info | | --------- | -------------------------------------------------------- | | Python | 3.10.19 (main, Oct 21 2025, 16:43:05) [GCC 11.2.0] | | OS | Linux-5.14.0-570.23.1.el9_6.x86_64-x86_64-with-glibc2.34 | | CPU | 32 logical CPU cores, x86_64 | | GPU | No GPU found | | Updated | 2026-01-20 19:17 |
Setting paths#
[3]:
path = '/home/mid166/Analysis/Jupyter/Python/Longitudinal/Integration'
import sys
sys.path.append(path)
from env_settings import *
sc.set_figure_params(dpi=80,dpi_save=300, color_map='viridis',facecolor='white')
rcParams['figure.figsize'] = 4, 4
save_dir='/n/scratch/users/m/mid166/Result/single-cell/Methods/COSG/Database'
### Create the save_dir if not existed
!mkdir -p {save_dir}
Load the adult mouse cortex RNA#
Google Drive link: https://drive.google.com/file/d/1bEyWNjGvoA9kz3J6jnbXRKgAIytb555W
mkdir -p /n/scratch/users/m/mid166/Result/single-cell/Enhancer/AdultCortexMultiome
cd /n/scratch/users/m/mid166/Result/single-cell/Enhancer/AdultCortexMultiome
gdrive files download 1bEyWNjGvoA9kz3J6jnbXRKgAIytb555W
[4]:
adata=sc.read('/n/scratch/users/m/mid166/Result/single-cell/Enhancer/AdultCortexMultiome/AdultCortexMultiomeRNA_integrated_anno.h5ad')
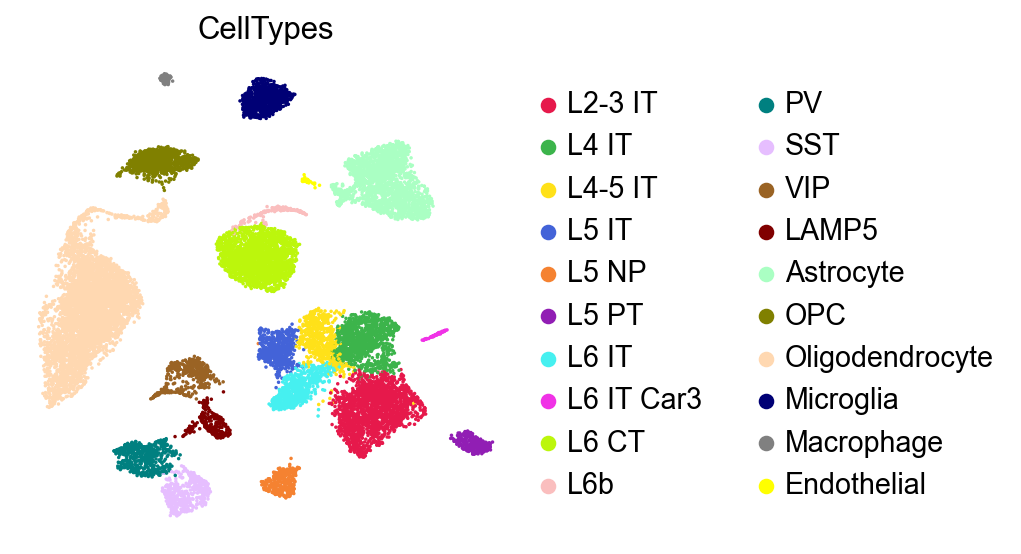
[5]:
sc.pl.umap(adata,
color=['CellTypes'],
palette=self_palette2,
# legend_loc='on data',
legend_fontoutline=2,
legend_fontweight=5,
cmap='Spectral_r',
ncols=3,
size=10,
frameon=False)
PIASOmarkerDB API#
PIASOmarkerDB is accessible via https://piaso.org/piasomarkerdb/.
Query by gene#
[6]:
marker_df = piaso.tl.queryPIASOmarkerDB(gene="Fezf2")
[7]:
marker_df.head(10)
[7]:
| cell_type | condition | gene | species | specificity_score | study_publication | tissue | |
|---|---|---|---|---|---|---|---|
| 0 | 119 SI-MA-LPO-LHA Skor1 Glut | normal | Fezf2 | Mouse | 6.785830 | AllenWholeMouseBrain_GABA | brain |
| 1 | 131 LHA-AHN-PVH Otp Trh Glut | normal | Fezf2 | Mouse | 6.384136 | AllenWholeMouseBrain_GABA | brain |
| 2 | 032 L5 NP CTX Glut | normal | Fezf2 | Mouse | 5.638977 | AllenWholeMouseBrain_Neuron | brain |
| 3 | Ex_Fezf2 | normal | Fezf2 | Mouse | 5.496620 | MacoskoWholeMouseBrain_nuclei_Neuron_AnnoL2 | brain |
| 4 | 023 SUB-ProS Glut | normal | Fezf2 | Mouse | 5.360841 | AllenWholeMouseBrain_Neuron | brain |
| 5 | 022 L5 ET CTX Glut | normal | Fezf2 | Mouse | 5.340233 | AllenWholeMouseBrain_Neuron | brain |
| 6 | 319 Astro-TE NN | normal | Fezf2 | Mouse | 5.327042 | AllenWholeMouseBrain_NonNeuron | brain |
| 7 | Ex_Vwc2l | normal | Fezf2 | Mouse | 4.739503 | MacoskoWholeMouseBrain_nuclei_Neuron_AnnoL2 | brain |
| 8 | SUB-ProS Glut_3 | normal; ageing | Fezf2 | Mouse | 4.675560 | AllenAgeingBrainAtlas2025_supertype | brain |
| 9 | 033 NP SUB Glut | normal | Fezf2 | Mouse | 4.618183 | AllenWholeMouseBrain_Neuron | brain |
Query by multiple genes#
[8]:
marker_df = piaso.tl.queryPIASOmarkerDB(gene=["Fezf2", "Satb2", "Tbr1"])
[9]:
marker_df.head(10)
[9]:
| cell_type | condition | gene | species | specificity_score | study_publication | tissue | |
|---|---|---|---|---|---|---|---|
| 0 | Fibro_Vdr | normal | Satb2 | Mouse | 13.416058 | MacoskoWholeMouseBrain_nuclei_NonNeuron_AnnoL2 | brain |
| 1 | 112 GPi Tbr1 Cngb3 Gaba-Glut | normal | Tbr1 | Mouse | 9.910396 | AllenWholeMouseBrain_GABA | brain |
| 2 | Ng_Lhx1 | normal | Tbr1 | Mouse | 9.585728 | MacoskoWholeMouseBrain_nuclei_NonNeuron_AnnoL2 | brain |
| 3 | 119 SI-MA-LPO-LHA Skor1 Glut | normal | Tbr1 | Mouse | 9.344679 | AllenWholeMouseBrain_GABA | brain |
| 4 | 088 BST Tac2 Gaba | normal | Satb2 | Mouse | 7.589609 | AllenWholeMouseBrain_GABA | brain |
| 5 | 099 SBPV-PVa Six6 Satb2 Gaba | normal | Satb2 | Mouse | 6.996956 | AllenWholeMouseBrain_GABA | brain |
| 6 | large intestine goblet cell | normal | SATB2 | Human | 6.796177 | TabulaSapiens_10X | multiple |
| 7 | enterocyte of epithelium of large intestine | normal | SATB2 | Human | 6.786366 | TabulaSapiens_10X | multiple |
| 8 | 119 SI-MA-LPO-LHA Skor1 Glut | normal | Fezf2 | Mouse | 6.785830 | AllenWholeMouseBrain_GABA | brain |
| 9 | 038 DG-PIR Ex IMN | normal | Tbr1 | Mouse | 6.649887 | AllenWholeMouseBrain_NonNeuron | brain |
Query by study#
[10]:
marker_df = piaso.tl.queryPIASOmarkerDB(
study="AllenWholeMouseBrain_isocortex",
species="mouse",
# limit=10
)
[11]:
marker_df
[11]:
| cell_type | condition | gene | species | specificity_score | study_publication | tissue | |
|---|---|---|---|---|---|---|---|
| 0 | 056 Sst Chodl Gaba | normal | Chodl | Mouse | 14.086876 | AllenWholeMouseBrain_isocortex | brain; cortex |
| 1 | 056 Sst Chodl Gaba | normal | Krt18 | Mouse | 13.081692 | AllenWholeMouseBrain_isocortex | brain; cortex |
| 2 | 051 Pvalb chandelier Gaba | normal | Vmn1r209 | Mouse | 12.826864 | AllenWholeMouseBrain_isocortex | brain; cortex |
| 3 | 330 VLMC NN | normal | Slc22a6 | Mouse | 11.932269 | AllenWholeMouseBrain_isocortex | brain; cortex |
| 4 | 335 BAM NN | normal | F13a1 | Mouse | 11.871344 | AllenWholeMouseBrain_isocortex | brain; cortex |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 1295 | 005 L5 IT CTX Glut | normal | Phex | Mouse | 1.709302 | AllenWholeMouseBrain_isocortex | brain; cortex |
| 1296 | 005 L5 IT CTX Glut | normal | Lynx1 | Mouse | 1.699085 | AllenWholeMouseBrain_isocortex | brain; cortex |
| 1297 | 005 L5 IT CTX Glut | normal | Gm2694 | Mouse | 1.677627 | AllenWholeMouseBrain_isocortex | brain; cortex |
| 1298 | 005 L5 IT CTX Glut | normal | A2ml1 | Mouse | 1.676827 | AllenWholeMouseBrain_isocortex | brain; cortex |
| 1299 | 005 L5 IT CTX Glut | normal | Gm50304 | Mouse | 1.671815 | AllenWholeMouseBrain_isocortex | brain; cortex |
1300 rows × 7 columns
Query with score filter#
[12]:
marker_df = piaso.tl.queryPIASOmarkerDB(
species="Human",
min_score=8.0,
limit=100
)
[13]:
marker_df.head(20)
[13]:
| cell_type | condition | gene | species | specificity_score | study_publication | tissue | |
|---|---|---|---|---|---|---|---|
| 0 | Neut | normal | CEACAM8 | Human | 22.576055 | YayonTeichmann2023Thymus_broad | thymus |
| 1 | Neut | normal | PGLYRP1 | Human | 22.467628 | YayonTeichmann2023Thymus_broad | thymus |
| 2 | Neut | normal | CAMP | Human | 22.386975 | YayonTeichmann2023Thymus_broad | thymus |
| 3 | Neut | normal | MMP8 | Human | 22.354010 | YayonTeichmann2023Thymus_broad | thymus |
| 4 | Neut | normal | TCN1 | Human | 22.239567 | YayonTeichmann2023Thymus_broad | thymus |
| 5 | Neut | normal | DEFA3 | Human | 22.141212 | YayonTeichmann2023Thymus_broad | thymus |
| 6 | Neut | normal | BPI | Human | 21.663849 | YayonTeichmann2023Thymus_broad | thymus |
| 7 | Myelocyte | normal | DEFA3 | Human | 21.135054 | YayonTeichmann2023Thymus | thymus |
| 8 | erythrocyte | normal | ALAS2 | Human | 21.131534 | TabulaSapiens_10X | multiple |
| 9 | erythrocyte | normal | ENSG00000290010.1 | Human | 21.073169 | TabulaSapiens_10X | multiple |
| 10 | erythrocyte | normal | HBM | Human | 20.988997 | TabulaSapiens_10X | multiple |
| 11 | Neut | normal | ARG1 | Human | 20.970731 | YayonTeichmann2023Thymus_broad | thymus |
| 12 | erythrocyte | normal | SLC4A1 | Human | 20.866775 | TabulaSapiens_10X | multiple |
| 13 | erythrocyte | normal | IFIT1B | Human | 20.742300 | TabulaSapiens_10X | multiple |
| 14 | erythrocyte | normal | HBQ1 | Human | 20.688987 | TabulaSapiens_10X | multiple |
| 15 | Neut | normal | CRISP3 | Human | 20.649712 | YayonTeichmann2023Thymus_broad | thymus |
| 16 | acinar cell | normal | LCN1P1 | Human | 20.612447 | TabulaSapiens_10X | multiple |
| 17 | erythrocyte | normal | ENSG00000290038.1 | Human | 20.544104 | TabulaSapiens_10X | multiple |
| 18 | erythrocyte | normal | EPB42 | Human | 20.501436 | TabulaSapiens_10X | multiple |
| 19 | erythrocyte | normal | AHSP | Human | 20.452516 | TabulaSapiens_10X | multiple |
Get Markers as Dictionary#
[14]:
marker_df, marker_dict = piaso.tl.queryPIASOmarkerDB(
study='AllenWholeMouseBrain_isocortex',
# species="Human",
# min_score=3.0,
# limit=500,
as_dict=True
)
[15]:
print(f"DataFrame shape: {marker_df.shape}")
print(f"Cell types in dict: {len(marker_dict)}")
# Show sample
for ct, genes in list(marker_dict.items())[:3]:
print(f"\n {ct}:")
print(f" Markers: {genes[:5]}...")
DataFrame shape: (1300, 7)
Cell types in dict: 26
056 Sst Chodl Gaba:
Markers: ['Chodl', 'Krt18', 'P2rx2', '4930545L08Rik', 'Tacr1']...
051 Pvalb chandelier Gaba:
Markers: ['Vmn1r209', 'Vmn1r207-ps', 'Dbhos', 'Nkx2-1', 'Sfta3-ps']...
330 VLMC NN:
Markers: ['Slc22a6', 'Aldh1a2', 'Slc13a4', 'Fam180a', 'Lum']...
List available studies#
[16]:
studies = piaso.tl.queryPIASOmarkerDB(list_studies=True)
print(f"Total studies: {len(studies)}")
print("First 10 studies:")
for study in studies[:10]:
print(f" - {study}")
Total studies: 36
First 10 studies:
- AllenAgeingBrainAtlas2025_supertype
- AllenDevVisualCortex2025_RNA
- AllenHumanImmuneHealthAtlas_L2
- AllenHumanImmuneHealthAtlas_L3
- AllenWholeMouseBrain_GABA
- AllenWholeMouseBrain_Neuron
- AllenWholeMouseBrain_NonNeuron
- AllenWholeMouseBrain_isocortex
- AsianImmuneDiversityAtlasPhase1v2_5prime
- DiBellaArlotta2021
Analyze single gene list#
[17]:
def example_analyze_single_list():
"""Demonstrate single gene list analysis."""
print("\n" + "="*60)
print("Example 4: Analyze Single Gene List")
print("="*60)
# T-cell marker genes
t_cell_genes = ["CD3E", "CD3D", "CD8A", "GZMK", "PRF1", "IFNG"]
print(f"\n--- Analyzing genes: {t_cell_genes} ---")
df = piaso.tl.analyzeMarkers(t_cell_genes, species="Human")
print(f"Found {len(df)} cell type matches")
if len(df) > 0:
print("\nTop 5 matches:")
print(df[['cell_type', 'matched_gene_count', 'avg_specificity']].head())
[18]:
query_genes = ["Syt6", "Tle4", "Hs3st4", "Col6a1", "Zfpm2", "Sema5a", "Bcl11b", "Fezf2", "Foxp2", "Col12a1"]
print(f"\n--- Analyzing genes: {query_genes} ---")
marker_df = piaso.tl.analyzeMarkers(query_genes)
marker_df.head()
--- Analyzing genes: ['Syt6', 'Tle4', 'Hs3st4', 'Col6a1', 'Zfpm2', 'Sema5a', 'Bcl11b', 'Fezf2', 'Foxp2', 'Col12a1'] ---
[18]:
| cell_type | study_publication | species | tissue | condition | matched_gene_count | matched_genes | avg_specificity | |
|---|---|---|---|---|---|---|---|---|
| 0 | EN-L6-CT | WangKriegstein2025 | Human | brain | normal | 6 | SYT6,HS3ST4,FOXP2,TLE4,FEZF2,ZFPM2 | 2.981403 |
| 1 | CThPN | DiBellaArlotta2021 | Mouse | brain; cortex | normal | 5 | Hs3st4,Syt6,Tle4,Zfpm2,Foxp2 | 4.958002 |
| 2 | 439_L6 CT CTX Glut_1 | AllenDevVisualCortex2025_RNA | Mouse | iscortex; brain | normal; developmental | 5 | Syt6,Foxp2,Tle4,Hs3st4,Zfpm2 | 3.561619 |
| 3 | 437_L6 CT CTX Glut_1 | AllenDevVisualCortex2025_RNA | Mouse | iscortex; brain | normal; developmental | 5 | Syt6,Foxp2,Zfpm2,Hs3st4,Tle4 | 2.928945 |
| 4 | 440_L6 CT CTX Glut_1 | AllenDevVisualCortex2025_RNA | Mouse | iscortex; brain | normal; developmental | 5 | Fezf2,Tle4,Col12a1,Hs3st4,Foxp2 | 2.678869 |
Analyze Multiple Gene Sets (Dictionary)#
[19]:
gene_sets = {
'Cluster_0': ['Cx3cr1', 'P2ry12', 'Teme119', 'Csf1r', 'Itgam', 'Aif1', 'Trem2'],
'Cluster_1': ["Syt6", "Tle4", "Hs3st4", "Col6a1", "Zfpm2", "Sema5a", "Bcl11b", "Fezf2", "Foxp2", "Col12a1"],
}
print("\n--- Input Gene Sets ---")
for name, genes in gene_sets.items():
print(f" {name}: {genes}")
print("\n--- Analyzing... ---")
# results, top_hits = piaso.tl.analyzeMarkers(gene_sets, species="Mouse")
results, top_hits = piaso.tl.analyzeMarkers(gene_sets)
print("\nCell Type Predictions:")
for cluster, cell_type in top_hits.items():
print(f" {cluster}: {cell_type}")
# Show detailed results
print(f"\n--- Detailed results for Cluster_0 ---")
if 'Cluster_0' in results and len(results['Cluster_0']) > 0:
print(results['Cluster_0'][['cell_type', 'matched_gene_count', 'avg_specificity', 'study_publication']].head())
print(f"\n--- Detailed results for Cluster_1 ---")
if 'Cluster_0' in results and len(results['Cluster_1']) > 0:
print(results['Cluster_1'][['cell_type', 'matched_gene_count', 'avg_specificity', 'study_publication']].head())
--- Input Gene Sets ---
Cluster_0: ['Cx3cr1', 'P2ry12', 'Teme119', 'Csf1r', 'Itgam', 'Aif1', 'Trem2']
Cluster_1: ['Syt6', 'Tle4', 'Hs3st4', 'Col6a1', 'Zfpm2', 'Sema5a', 'Bcl11b', 'Fezf2', 'Foxp2', 'Col12a1']
--- Analyzing... ---
Cell Type Predictions:
Cluster_0: Microglia
Cluster_1: EN-L6-CT
--- Detailed results for Cluster_0 ---
cell_type matched_gene_count avg_specificity \
0 Microglia 5 13.000120
1 Mgl_9 5 7.875872
2 5312_Microglia NN_1 4 8.265331
3 P2RY12+ microglia 4 7.782557
4 Mgl_11 4 7.375123
study_publication
0 DiBellaArlotta2021
1 SilettiLinnarssonWholeHumanBrain2023_subtype
2 AllenDevVisualCortex2025_RNA
3 XuTeichmann2023_HippocampalFormation
4 SilettiLinnarssonWholeHumanBrain2023_subtype
--- Detailed results for Cluster_1 ---
cell_type matched_gene_count avg_specificity \
0 EN-L6-CT 6 2.981403
1 CThPN 5 4.958002
2 439_L6 CT CTX Glut_1 5 3.561619
3 437_L6 CT CTX Glut_1 5 2.928945
4 440_L6 CT CTX Glut_1 5 2.678869
study_publication
0 WangKriegstein2025
1 DiBellaArlotta2021
2 AllenDevVisualCortex2025_RNA
3 AllenDevVisualCortex2025_RNA
4 AllenDevVisualCortex2025_RNA
COSG integration#
The piaso.tl.analyzeMarkers function will infer the cell types for each cluster or cell group based on the top marker genes identified by the marker gene identification method COSG and the cell type marker genes collected in PIASOmarkerDB.
Run COSG#
[20]:
import cosg
You could set groupby to Leiden, here is using the manually annotated cell type column as a demo:
[21]:
%%time
groupby='CellTypes'
cosg.cosg(adata,
key_added='cosg',
use_raw=False, layer='log1p', ## e.g., if you want to use the log1p layer in adata
mu=100,
expressed_pct=0.1,
remove_lowly_expressed=True,
n_genes_user=adata.n_vars, ### Use all the genes, to enable the calculation of transformed COSG scores
# n_genes_user=100,
groupby=groupby,
return_by_group=True,
verbosity=1
)
Finished identifying marker genes by COSG, and the results are in adata.uns['cosg'].
CPU times: user 2.54 s, sys: 366 ms, total: 2.9 s
Wall time: 2.91 s
Here, we take the top 50 marker genes for each group:
[22]:
cosg_marker_df=pd.DataFrame(adata.uns['cosg']['names']).head(50)
[23]:
cosg_marker_df.head()
[23]:
| L2-3 IT | L4 IT | L4-5 IT | L5 IT | L5 NP | L5 PT | L6 IT | L6 IT Car3 | L6 CT | L6b | PV | SST | VIP | LAMP5 | Astrocyte | OPC | Oligodendrocyte | Microglia | Macrophage | Endothelial | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Otof | Rspo1 | Scn7a | Deptor | Trhr | Gm42705 | Slc26a4 | Gm6602 | Foxp2 | Nxph4 | Adamts15 | Sst | Vip | Ndnf | Gja1 | Pdgfra | St18 | Selplg | F13a1 | Slco1a4 |
| 1 | Ccbe1 | Gm40331 | Tnnc1 | Il1rapl2 | Myzap | Shoc1 | Bmp3 | Car3 | Col5a1 | Ccn2 | Pvalb | Pdyn | Htr3a | Pde11a | Gli3 | Cspg4 | Prr5l | Siglech | Ms4a4a | Cldn5 |
| 2 | Evc2 | Gm42953 | BC006965 | Rxfp2 | Slc17a8 | Gm42707 | Dnah14 | Gm49272 | Syt6 | Gm41414 | 6330411D24Rik | Hpse | Tac2 | Ltbp2 | Gjb6 | C1ql1 | Mog | Tmem119 | Pf4 | Adgrl4 |
| 3 | B230216N24Rik | Cbln4 | Hgf | Fras1 | Mirt1 | L3mbtl4 | Rspo2 | Oprk1 | Cpa6 | Cplx3 | Tac1 | Tnni3k | Grpr | Myo3a | Ntsr2 | Tmem255b | Cldn11 | Gm2629 | Cd163 | Flt1 |
| 4 | Efcab1 | Col8a1 | 1700047F07Rik | Tmem91 | Hmga2 | Gm6260 | Galnt14 | BB557941 | Rprm | Clic5 | Gm26633 | Crhbp | Npy2r | Myo3b | Gm6145 | Neu4 | Mobp | Olfml3 | Aoah | Abcb1a |
[24]:
%%time
results, top_hits = piaso.tl.analyzeMarkers(
cosg_marker_df,
n_top_genes=50,
species="mouse",
)
CPU times: user 576 ms, sys: 19.5 ms, total: 596 ms
Wall time: 3.56 s
[25]:
print("\nCell Type Predictions:")
for cluster, cell_type in top_hits.items():
print(f" {cluster}: {cell_type}")
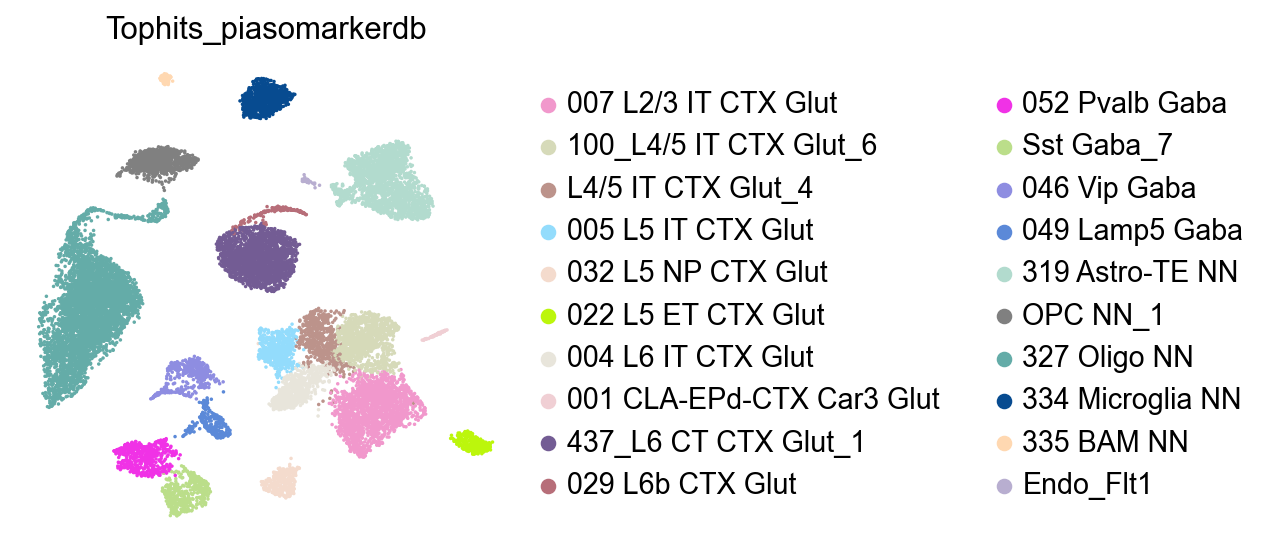
Cell Type Predictions:
L2-3 IT: 007 L2/3 IT CTX Glut
L4 IT: 100_L4/5 IT CTX Glut_6
L4-5 IT: L4/5 IT CTX Glut_4
L5 IT: 005 L5 IT CTX Glut
L5 NP: 032 L5 NP CTX Glut
L5 PT: 022 L5 ET CTX Glut
L6 IT: 004 L6 IT CTX Glut
L6 IT Car3: 001 CLA-EPd-CTX Car3 Glut
L6 CT: 437_L6 CT CTX Glut_1
L6b: 029 L6b CTX Glut
PV: 052 Pvalb Gaba
SST: Sst Gaba_7
VIP: 046 Vip Gaba
LAMP5: 049 Lamp5 Gaba
Astrocyte: 319 Astro-TE NN
OPC: OPC NN_1
Oligodendrocyte: 327 Oligo NN
Microglia: 334 Microglia NN
Macrophage: 335 BAM NN
Endothelial: Endo_Flt1
[26]:
results['L2-3 IT'].head(5)
[26]:
| cell_type | study_publication | species | tissue | condition | matched_gene_count | matched_genes | avg_specificity | |
|---|---|---|---|---|---|---|---|---|
| 0 | 007 L2/3 IT CTX Glut | AllenWholeMouseBrain_isocortex | Mouse | brain; cortex | normal | 22 | Otof,Efcab1,Gm10754,Evc2,Ccbe1,B230216N24Rik,F... | 3.148077 |
| 1 | 007 L2/3 IT CTX Glut | AllenWholeMouseBrain_Neuron | Mouse | brain | normal | 16 | Gm48530,Evc2,Igfn1,A830009L08Rik,Lamp5,Gm20063... | 3.244823 |
| 2 | L2/3 IT CTX Glut_2 | AllenAgeingBrainAtlas2025_supertype | Mouse | brain | normal; ageing | 11 | Clec18a,Igfn1,Ccbe1,Gm48530,Lamp5,Gm20063,E130... | 3.554956 |
| 3 | L2/3 IT CTX Glut_1 | AllenAgeingBrainAtlas2025_supertype | Mouse | brain | normal; ageing | 11 | Ccbe1,Gm48530,Otof,Itga8,Evc2,E130304I02Rik,Du... | 3.502616 |
| 4 | 110_L2/3 IT CTX Glut_2 | AllenDevVisualCortex2025_RNA | Mouse | iscortex; brain | normal; developmental | 11 | Glra3,Ccbe1,Ccn3,6530403H02Rik,Gm42722,Greb1l,... | 2.801682 |
Check the prediction results#
Add annotations to AnnData:
[27]:
adata.obs['Tophits_piasomarkerdb']=adata.obs['CellTypes'].map(top_hits)
[28]:
sc.pl.umap(adata,
color=['Tophits_piasomarkerdb'],
palette=piaso.pl.color.d_color10,
# legend_loc='on data',
legend_fontoutline=2,
legend_fontweight=5,
cmap='Spectral_r',
ncols=3,
size=10,
frameon=False)
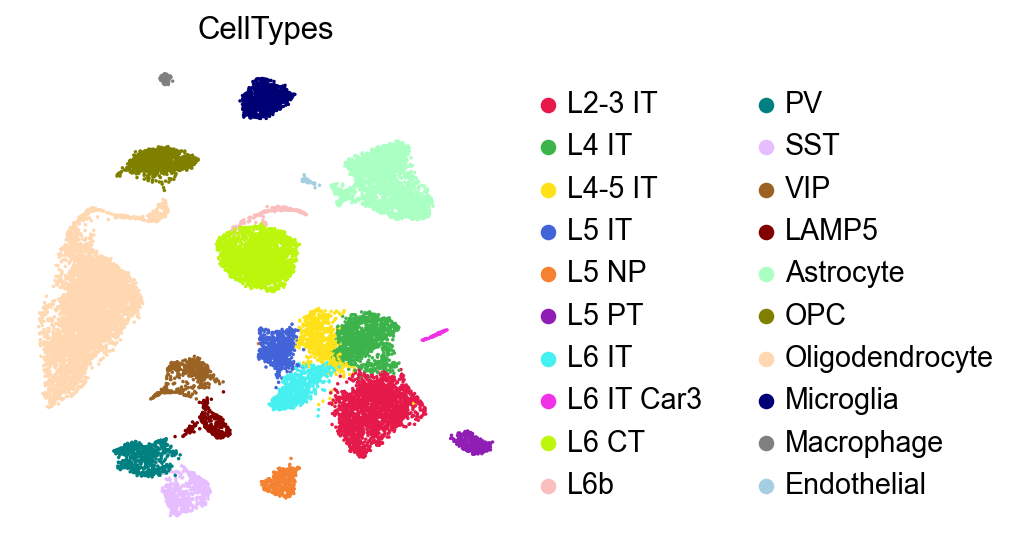
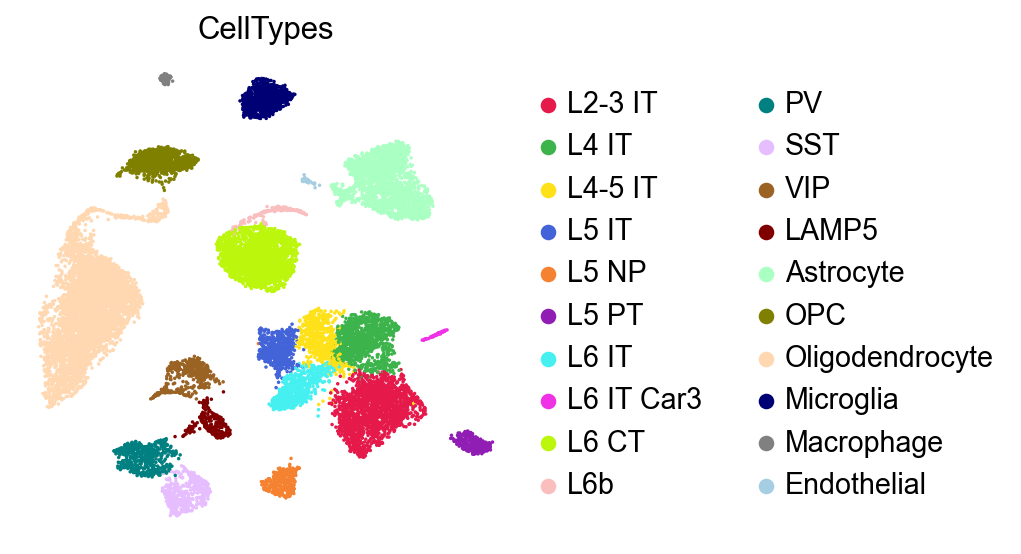
The mannual annotation:
[29]:
sc.pl.umap(adata,
color=['CellTypes'],
palette=piaso.pl.color.d_color4,
# legend_loc='on data',
legend_fontoutline=2,
legend_fontweight=5,
cmap='Spectral_r',
ncols=3,
size=10,
frameon=False)
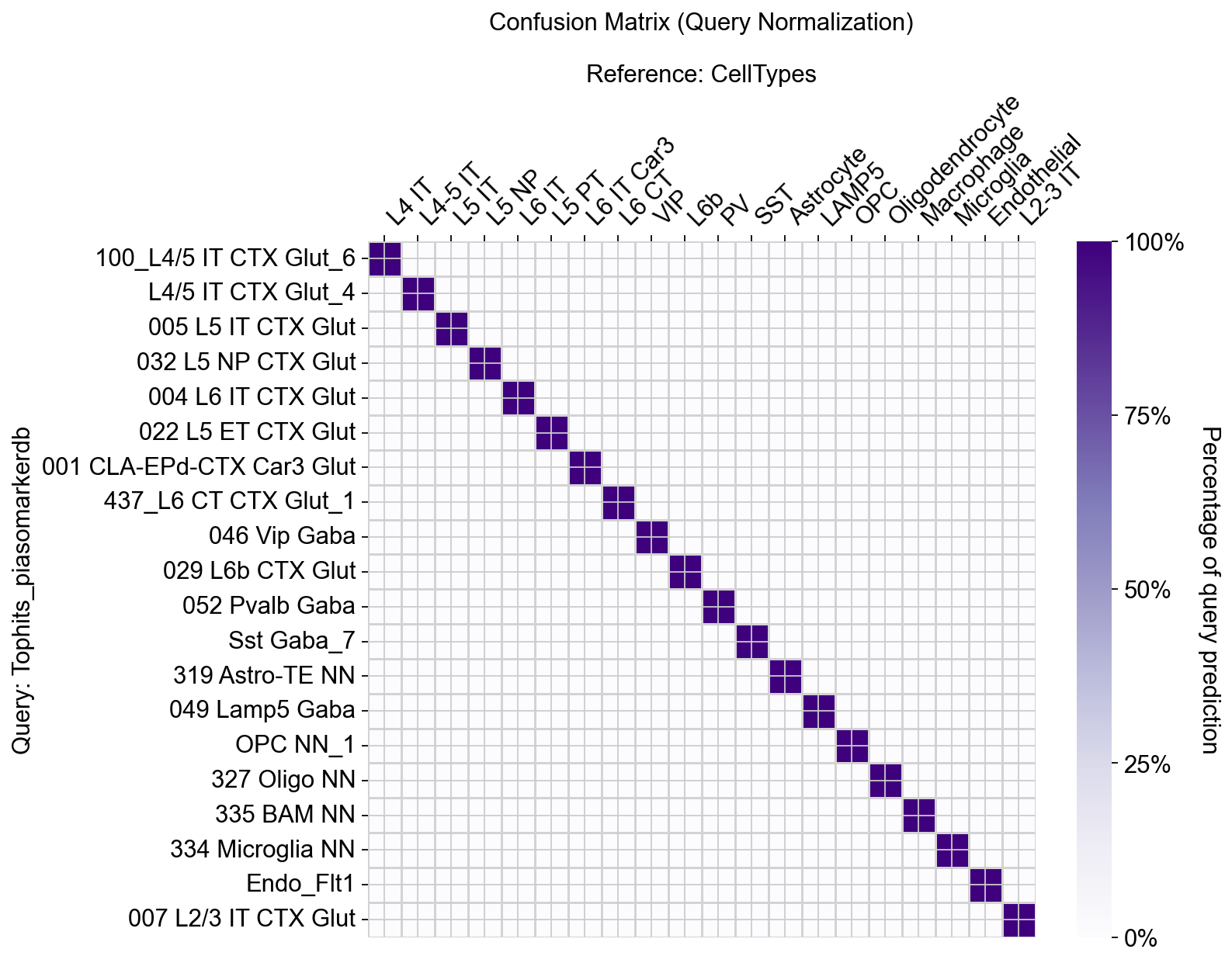
[30]:
piaso.pl.plotConfusionMatrix(adata, 'Tophits_piasomarkerdb', 'CellTypes', figsize=(10, 8))
Specify which study or studies to use in piaso.tl.analyzeMarkers#
[31]:
%%time
results, top_hits = piaso.tl.analyzeMarkers(
cosg_marker_df,
n_top_genes=50,
min_genes=5,
studies=['AllenWholeMouseBrain_isocortex'],
species="mouse"
)
CPU times: user 461 ms, sys: 7.07 ms, total: 469 ms
Wall time: 3.58 s
[32]:
top_hits
[32]:
{'L2-3 IT': '007 L2/3 IT CTX Glut',
'L4 IT': '006 L4/5 IT CTX Glut',
'L4-5 IT': '006 L4/5 IT CTX Glut',
'L5 IT': '005 L5 IT CTX Glut',
'L5 NP': '032 L5 NP CTX Glut',
'L5 PT': '022 L5 ET CTX Glut',
'L6 IT': '004 L6 IT CTX Glut',
'L6 IT Car3': '001 CLA-EPd-CTX Car3 Glut',
'L6 CT': '030 L6 CT CTX Glut',
'L6b': '029 L6b CTX Glut',
'PV': '052 Pvalb Gaba',
'SST': '053 Sst Gaba',
'VIP': '046 Vip Gaba',
'LAMP5': '049 Lamp5 Gaba',
'Astrocyte': '319 Astro-TE NN',
'OPC': '326 OPC NN',
'Oligodendrocyte': '327 Oligo NN',
'Microglia': '334 Microglia NN',
'Macrophage': '335 BAM NN',
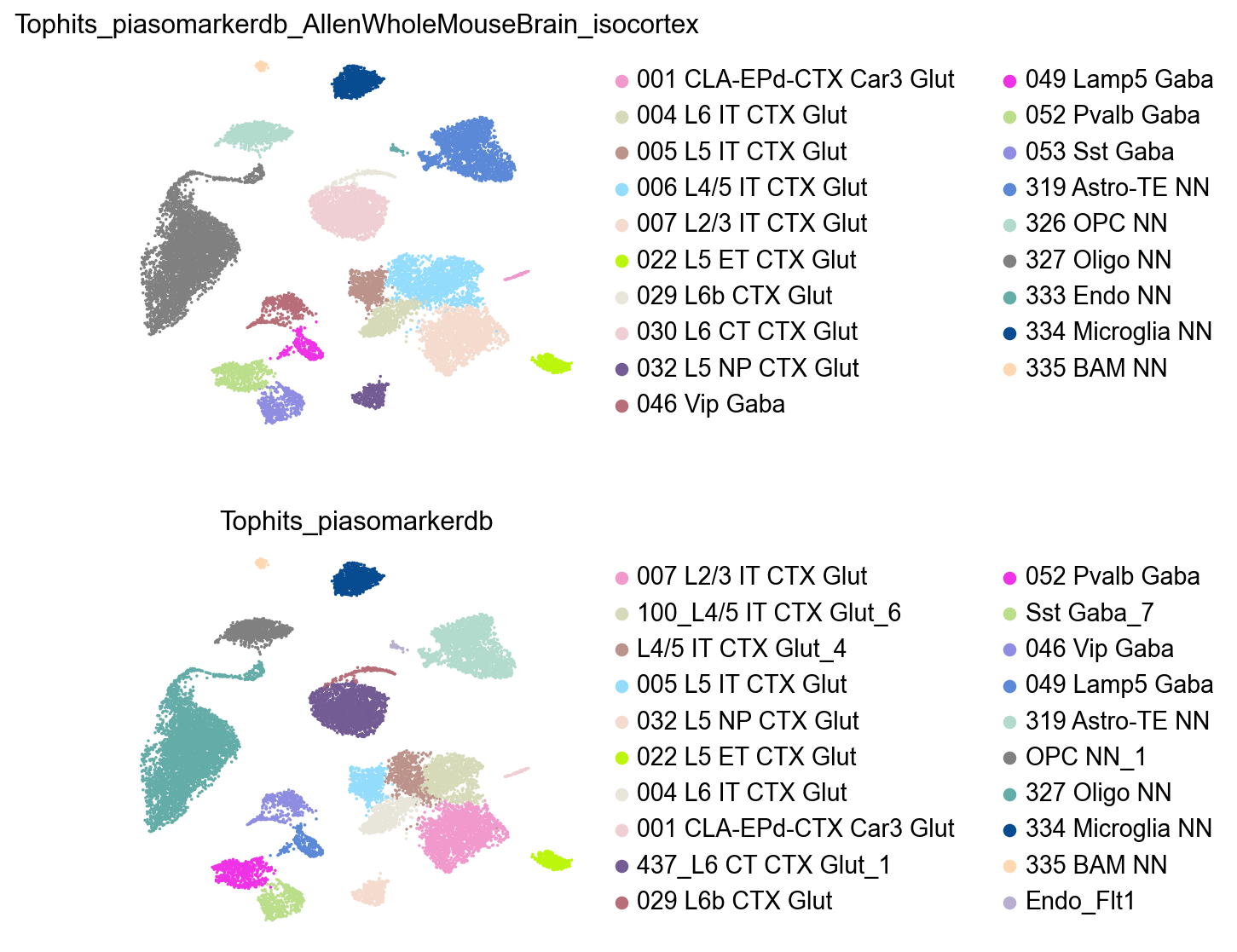
'Endothelial': '333 Endo NN'}
Add annotations to AnnData:
[33]:
adata.obs['Tophits_piasomarkerdb_AllenWholeMouseBrain_isocortex']=adata.obs['CellTypes'].map(top_hits)
We can see the predicted annotation on the L4/5 IT is different, it’s because in the AllenWholeMouseBrain_isocortex study’s annotation, the L4/5 IT and L4 IT are not distinguished at that annotation resolution:
[34]:
sc.pl.umap(adata,
color=[
'Tophits_piasomarkerdb_AllenWholeMouseBrain_isocortex',
'Tophits_piasomarkerdb'
],
palette=piaso.pl.color.d_color10,
# legend_loc='on data',
legend_fontoutline=2,
legend_fontweight=5,
cmap='Spectral_r',
ncols=1,
size=10,
frameon=False)
The mannual annotation:
[35]:
sc.pl.umap(adata,
color=['CellTypes'],
palette=piaso.pl.color.d_color4,
# legend_loc='on data',
legend_fontoutline=2,
legend_fontweight=5,
cmap='Spectral_r',
ncols=3,
size=10,
frameon=False)
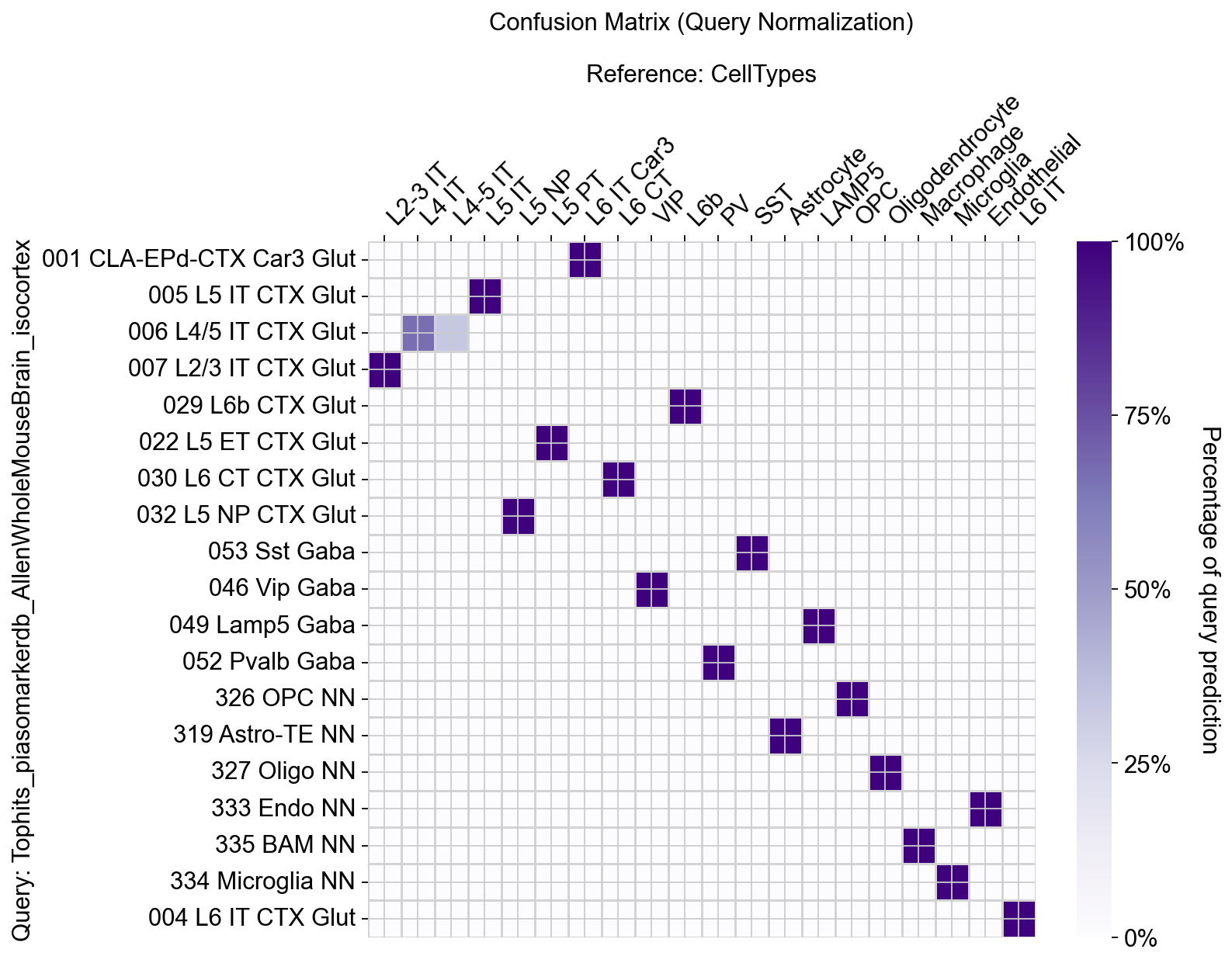
[36]:
piaso.pl.plotConfusionMatrix(adata, 'Tophits_piasomarkerdb_AllenWholeMouseBrain_isocortex', 'CellTypes', figsize=(10, 8))
Client Usage#
[37]:
client = piaso.tl.PIASOmarkerDB()
print(f"\nClient: {client}")
Client: PIASOmarkerDB(base_url='https://piaso.org/piasomarkerdb')
[38]:
# Get all markers with pagination
print("\n--- Get all markers for a gene (with pagination) ---")
df = client.getAllMarkers(gene="Chrna2", verbose=True)
print(f"Total Chrna2 entries: {len(df)}")
df.head(10)
--- Get all markers for a gene (with pagination) ---
Fetching markers... batch 1 (32 records)
Total: 32 markers
Total Chrna2 entries: 32
[38]:
| cell_type | condition | gene | species | specificity_score | study_publication | tissue | |
|---|---|---|---|---|---|---|---|
| 0 | 214 IPN Otp Crisp1 Gaba | normal | Chrna2 | Mouse | 11.105208 | AllenWholeMouseBrain_Neuron | brain |
| 1 | 214 IPN Otp Crisp1 Gaba | normal | Chrna2 | Mouse | 9.941944 | AllenWholeMouseBrain_GABA | brain |
| 2 | IPN Otp Crisp1 Gaba_2 | normal; ageing | Chrna2 | Mouse | 9.447727 | AllenAgeingBrainAtlas2025_supertype | brain |
| 3 | Inh_Lmo2 | normal | Chrna2 | Mouse | 9.027516 | MacoskoWholeMouseBrain_nuclei_Neuron_AnnoL2 | brain |
| 4 | VIP InN | normal | CHRNA2 | Human | 8.613732 | XuTeichmann2023_HippocampalFormation | hippocampal formation |
| 5 | IN-CGE-VIP | normal | CHRNA2 | Human | 7.967901 | WangKriegstein2025 | brain |
| 6 | Vip | normal; Alzheimer's disease | CHRNA2 | Human | 7.771841 | SEAAD2024_MTG_Subclass | brain; cortex |
| 7 | 779_Sst Gaba_4 | normal; developmental | Chrna2 | Mouse | 7.725675 | AllenDevVisualCortex2025_RNA | iscortex; brain |
| 8 | 780_Sst Gaba_4 | normal; developmental | Chrna2 | Mouse | 7.495388 | AllenDevVisualCortex2025_RNA | iscortex; brain |
| 9 | IPN Otp Crisp1 Gaba_1 | normal; ageing | Chrna2 | Mouse | 7.416458 | AllenAgeingBrainAtlas2025_supertype | brain |
[39]:
# Get recommended study
print("\n--- Recommended Studies ---")
for species, tissue in [("human", "blood"), ("human", "brain"), ("human", "spleen")]:
study = client.getRecommendedStudy(species, tissue)
print(f" {species}/{tissue}: {study}")
--- Recommended Studies ---
human/blood: AllenHumanImmuneHealthAtlas_L2
human/brain: SilettiLinnarssonWholeHumanBrain2023_class
human/spleen: XuTeichmann2023_Spleen
Invitation code to download from PIASOmarkerDB#
The invitation code is PIASO121125, and the code will be used during the beta testing phase and will be removed at later stages.
